6 Reference populations analysis
In this chapter, we will see how to compare the uploaded populations to reference populations (based on allele frequencies).
6.1 MDS on reference frequencies
STRAF implements an MDS computation based on allele frequency data. By default, allele frequencies from the STRiDER database (Bodner et al. 2016) are used (loci with less than 10 populations have been excluded). If you are not familiar with the MDS method and interpretation, you can find details in Chapter 4.
You can see the output of the MDS in the Reference population tab. It includes the MDS projection in two dimensions for 19 European populations, as well as the population tree.
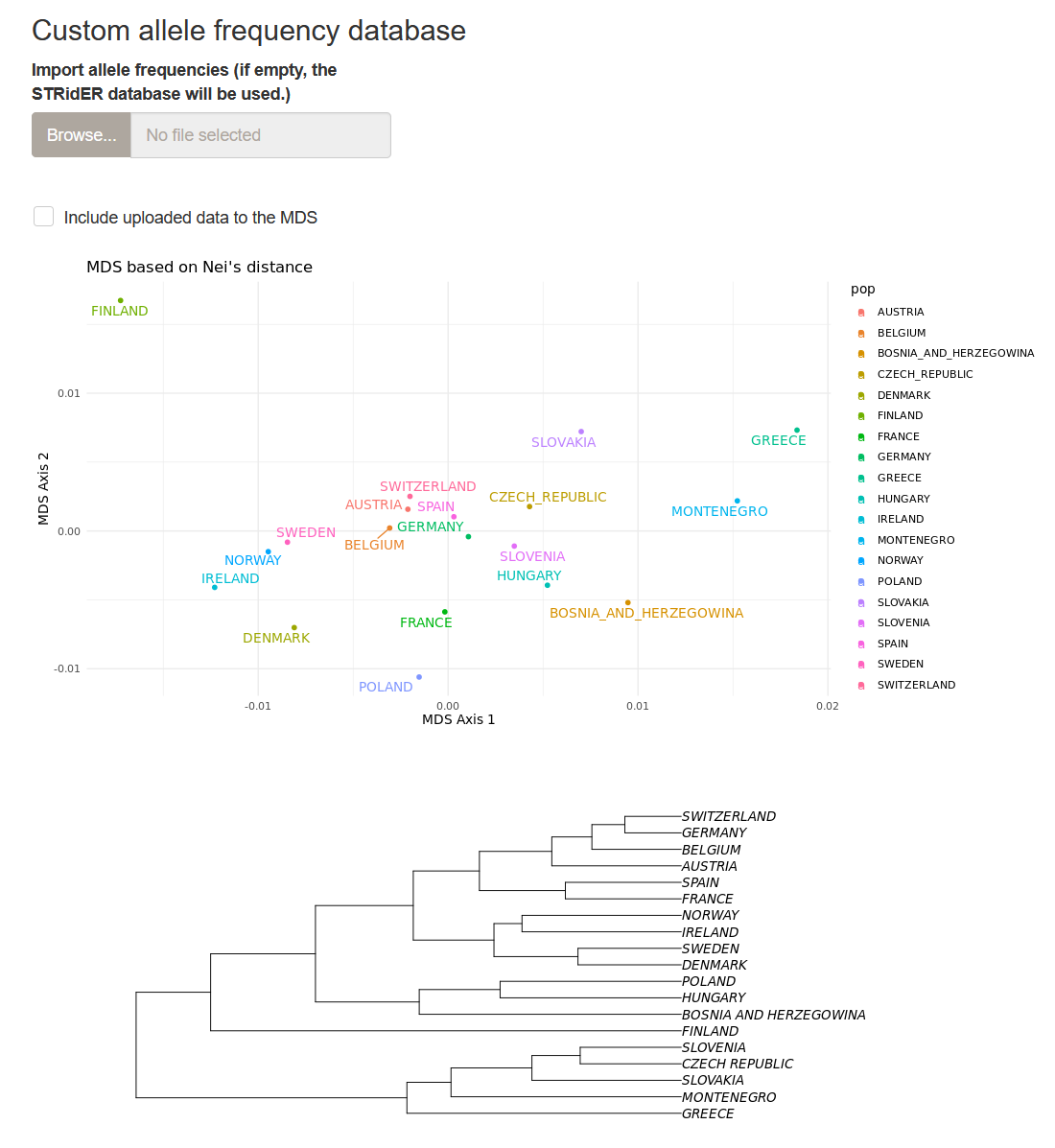
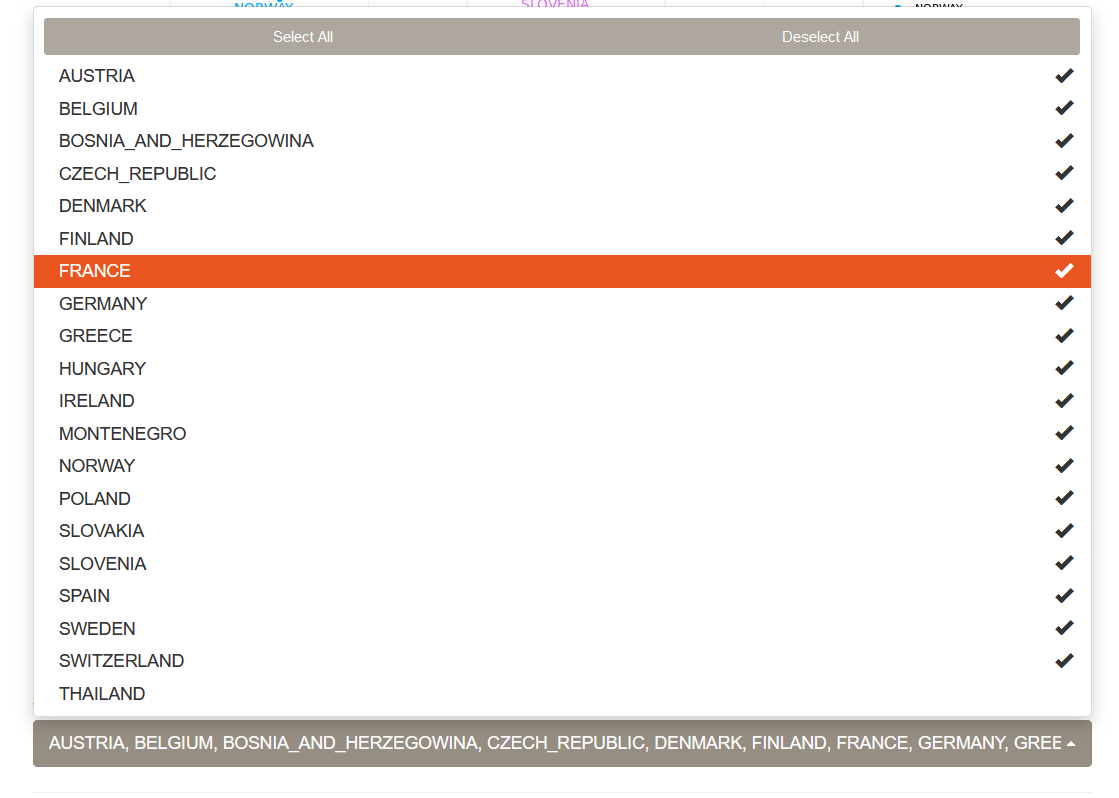
The drop-down menu below the plots allows one to select or unselect populations to be used in the MDS analysis.
Provided that some loci are in common between the reference allele frequencies and the genotypes you uploaded, it is possible to add your own populations to the existing MDS by checking the Include uploaded data to the MDS box.
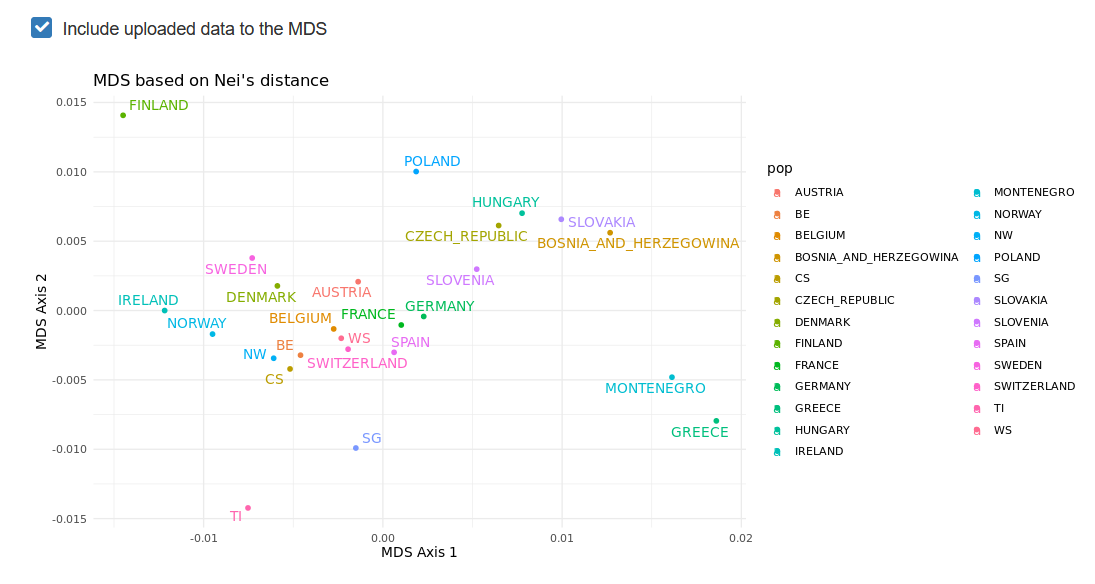
You will then see your populations added to the MDS plots.
6.2 Preparing a custom allele frequency database
If you want to use other reference databases than STRIDER, for example if you’re working with non-European samples or with different loci, it is possible to upload a custom allele frequency database in STRAF.
The data must be formatted as follows:
| D1S1656 | |||
|---|---|---|---|
| Allele | Switzerland | Germany | France |
| 9 | 0.12 | 0.09 | 0 |
| 10 | 0.40 | 0.35 | 0.28 |
| 10.2 | 0.31 | 0.41 | 0.5 |
| 11 | 0.17 | 0.15 | 0.22 |
| D2S1338 | |||
| Allele | Switzerland | Germany | France |
| 19 | 0.40 | 0.38 | 0.42 |
| 20 | 0.29 | 0.26 | 0.28 |
| 21 | 0.31 | 0.36 | 0.3 |
Starting from an Excel file for example, we can start with a spreadsheet that looks like this:
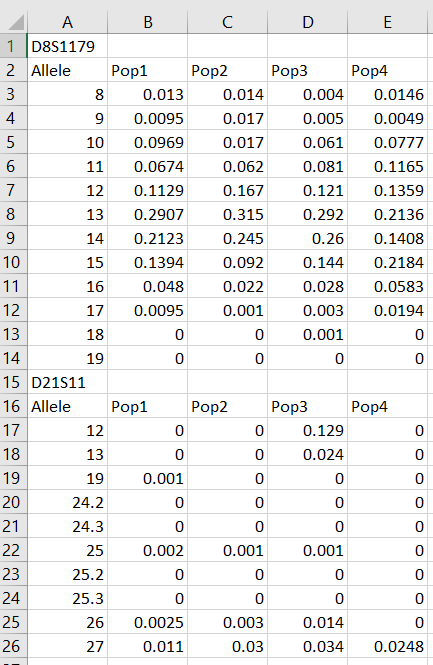
Then, one simply needs to save this table as a CSV (Comma-Separated Values) file. This can be
achieved by clicking on Save As > CSV (Comma-delimited) (*.csv).
Then, you can upload your data and use it as described for the STRIDER database.